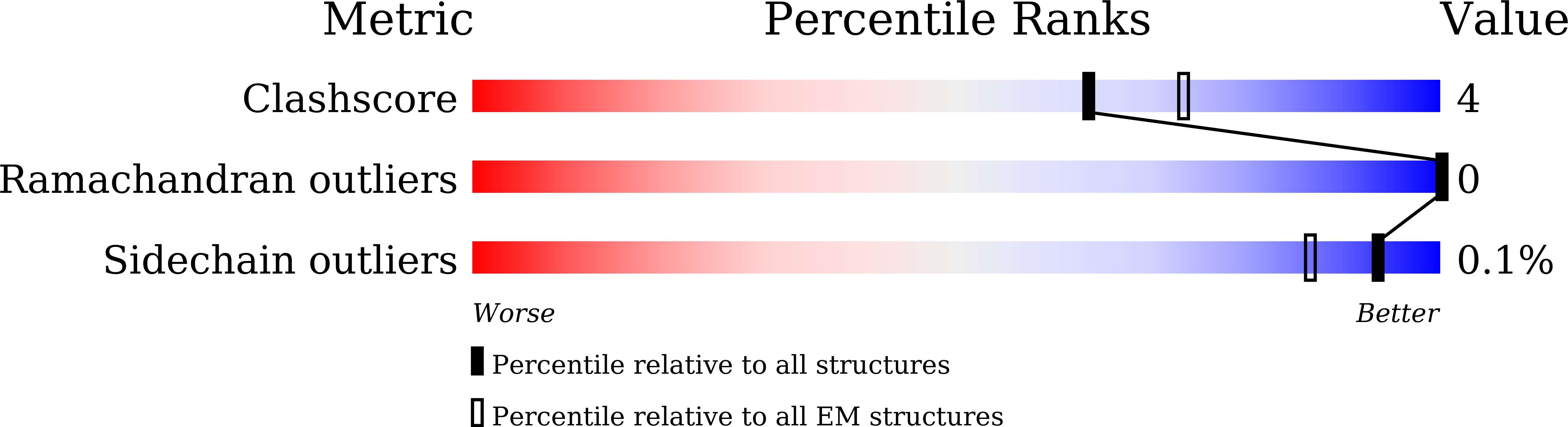Structural insights into hormone recognition by the human glucose-dependent insulinotropic polypeptide receptor.
Zhao, F., Zhang, C., Zhou, Q., Hang, K., Zou, X., Chen, Y., Wu, F., Rao, Q., Dai, A., Yin, W., Shen, D.D., Zhang, Y., Xia, T., Stevens, R.C., Xu, H.E., Yang, D., Zhao, L., Wang, M.W.(2021) Elife 10
- PubMed: 34254582
- DOI: https://doi.org/10.7554/eLife.68719
- Primary Citation of Related Structures:
7DTY - PubMed Abstract:
Glucose-dependent insulinotropic polypeptide (GIP) is a peptide hormone that exerts crucial metabolic functions by binding and activating its cognate receptor, GIPR. As an important therapeutic target, GIPR has been subjected to intensive structural studies without success. Here, we report the cryo-EM structure of the human GIPR in complex with GIP and a G s heterotrimer at a global resolution of 2.9 Å. GIP adopts a single straight helix with its N terminus dipped into the receptor transmembrane domain (TMD), while the C terminus is closely associated with the extracellular domain and extracellular loop 1. GIPR employs conserved residues in the lower half of the TMD pocket to recognize the common segments shared by GIP homologous peptides, while uses non-conserved residues in the upper half of the TMD pocket to interact with residues specific for GIP. These results provide a structural framework of hormone recognition and GIPR activation.
Organizational Affiliation:
School of Pharmacy, Fudan University, Shanghai, China.